ImmunoAnalysis. 4:4.
doi: 10.34172/ia.4070
Review
SARS-CoV-2 Mutations and Their Impacts on Virulence and Transmissibility of The Virus
Sajjad Bahman Conceptualization, Investigation, Writing – original draft, Writing – review & editing, 1 
Safar Farajnia Methodology, Project administration, Supervision, Validation, 2, 3, * 
Effat Alizadeh Formal analysis, 1
Hojjatollah Nozad Charoudeh Formal analysis, 4
Mohammad Kazem Hosseini Resources, 5
Behrooz Naghili Validation, 6
Author information:
1Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
2Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
3Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
4Stem Cell Research Center and Applied Cell Sciences Department, Tabriz University of Medical Sciences, Tabriz, Iran
5Istanbul University, Faculty of Sciences, Molecular Biology and Genetics Department, Istanbul, Turkey
6Infectious and Tropical Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
The emergence of SARS-CoV-2 in 2019 and its rapid global spread led to infrequent problems for human society. This vast spreading and high mortality rate were related mainly to the potential of this virus to develop mutated variants. Due to the emergence of hyper mutated strains escaping from vaccine-induced immune responses, the pandemic is still ongoing despite the discovery of various vaccines, and there is little chance that it will be eradicated in the near future. Therefore, it is crucial to monitor covid-19 mutations globally in order to develop new control measures. The SARS-CoV-2 mutation’s emergence and molecular characteristics have been the major talking points of this article, with a focus on significant variants classified as variants of concern (VOC).
Keywords: SARS-CoV-2, COVID-19, Mutation, Variant of concern, Omicron
Copyright and License Information
© 2024 The Author(s).
This is an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Funding Statement
This work was supported by the Biotechnology Research Center, Tabriz University of Medical Sciences (Grant no. 68941).
Introduction
With the emergence of the novel coronavirus in 2019, the world has faced a new pandemic disease called COVID-19. Due to its resemblance to the coronavirus that causes severe acute respiratory syndrome (SARS-CoV), a new member of the Coronaviridae family has been named SARS-CoV-2 by the International Committee on Taxonomy of Viruses.1 Similar to other coronaviruses, SARS-CoV-2 is an enveloped positive-sense virus. Fusion of the virus with the host membrane and penetration into the host cell is assisted by the envelope structure.2 The virus enters the host cell by adhering the viral spike to the receptor of angiotensin-converting enzyme 2 (ACE-2) on the surface of the host cell membrane.3
The timeline of SARS-CoV-2 spreading worldwide, divided into parts by CDC. First part was “Late 2019” which the WHO country office in China has received reports of an unidentified cause of pneumonia in Wuhan, China, presenting symptoms such as breathlessness and fever. Second part was in “Early 2020” that the CDC shares information about the outbreak of the 2019 novel coronavirus (2019-nCoV) caused by the SARS CoV-2 virus on its website. By “Late 2020” the second wave of COVID-19 accrued and in “Early 2021”, some countries experienced a third wave of COVID-19 cases. These waves were often associated with the emergence of new variants of the virus, which were more transmissible and sometimes exhibited increased disease severity. As of 22 May 2023, statistics from the WHO website show that SARS-CoV-2 has infected more than 766 million individuals and caused more than 6.9 million fatalities globally.
This virus is distinguished from other coronaviruses by the occurrence of successive mutations, which lead to increased pathogenicity and disease transmission. The effectiveness of vaccines, treatment strategies, and diagnostic methods may be affected by the recent global emergence of novel SARS-CoV-2 mutated variants. In addition, some of these mutations have increased the virus spreading rate and could make it more contagious and deadly.4 Therefore, it is important to continuously monitor viral mutations and their impact on pathogenicity, virulence, and transmission of the virus.
This article is dedicated to the investigation and follow-up of SARS-CoV-2 mutations and their spread worldwide. Also, effects of mutations on virus structure, immunogenicity, and transmission have been discussed.
Materials and Methods
Structure of SARS-CoV2 spike antigen
The Spike protein contains S1 and S2 domains, in which the S1 region consists of RBD and NTD subdomains. These areas are immunogenic and essential for viral attachment and cell entry.5-7 Most SARS-CoV-2 vaccines target this region of the S-glycoprotein. Many reported mutations are detected in glycoprotein S and affect the manner in which the virus attaches to host cells.8 These mutations enhance the likelihood of infection or reinfection, decrease the potency of vaccination, and allow the virus to escape neutralizing antibodies.
Nomenclature of mutations
SARS-CoV-2 has been classified using various nomenclature systems, including Pango, NextStrain, GISAID, and the Greek alphabet. The Greek and Pangolin letters were used in this manuscript for mentioning different variants. The Pangolin nomenclature, which stands for the Phylogenetic Assignment of Named Global Outbreak Lineages, was created to apply the dynamic naming of SARS-CoV-2 lineages.9,10
Results and Discussion
Origin, emergence, and dissemination of SARS-CoV-2 mutations
According to previous studies, shortly after the initiation of the pandemic, new strains emerged from the primary Wuhan-Hu-1 wild-type variant of SARS-CoV-2, each containing mutations that were correlated with the variant’s death rate, transmissibility, immune evasion, and resistance to neutralizing antibodies.
The first important mutant was the Alpha (B.1.1.7) (Figure 1a) strainreported in the United Kingdom in December 2020. It then spread worldwide and became the dominant variant in most patients; the WHO classified this mutant as a variant of concern (VOC).11,12 It has 23 mutations compared with the Wuhan-Hu-1 wild-type strain.13 By December 2020, Beta (B.1.351) (Figure 1b) with 13 mutations had been documented in South Africa.13 In October 2020, Delta (B.1.617.2) (Figure 1c), a new VOC, was reported in India. It is more contagious in comparison with the initial variant of SARS-CoV-2 and spreads very fast worldwide.13-16 In January 2021, the National Institute of Infectious Diseases of Japan reported a new variant of SARS-CoV-2 isolated from passengers who had traveled to Brazil; thus, the WHO labeled Gamma (P.1) (Figure 1d). It contains 17 mutationsand is considered VOC.13,17
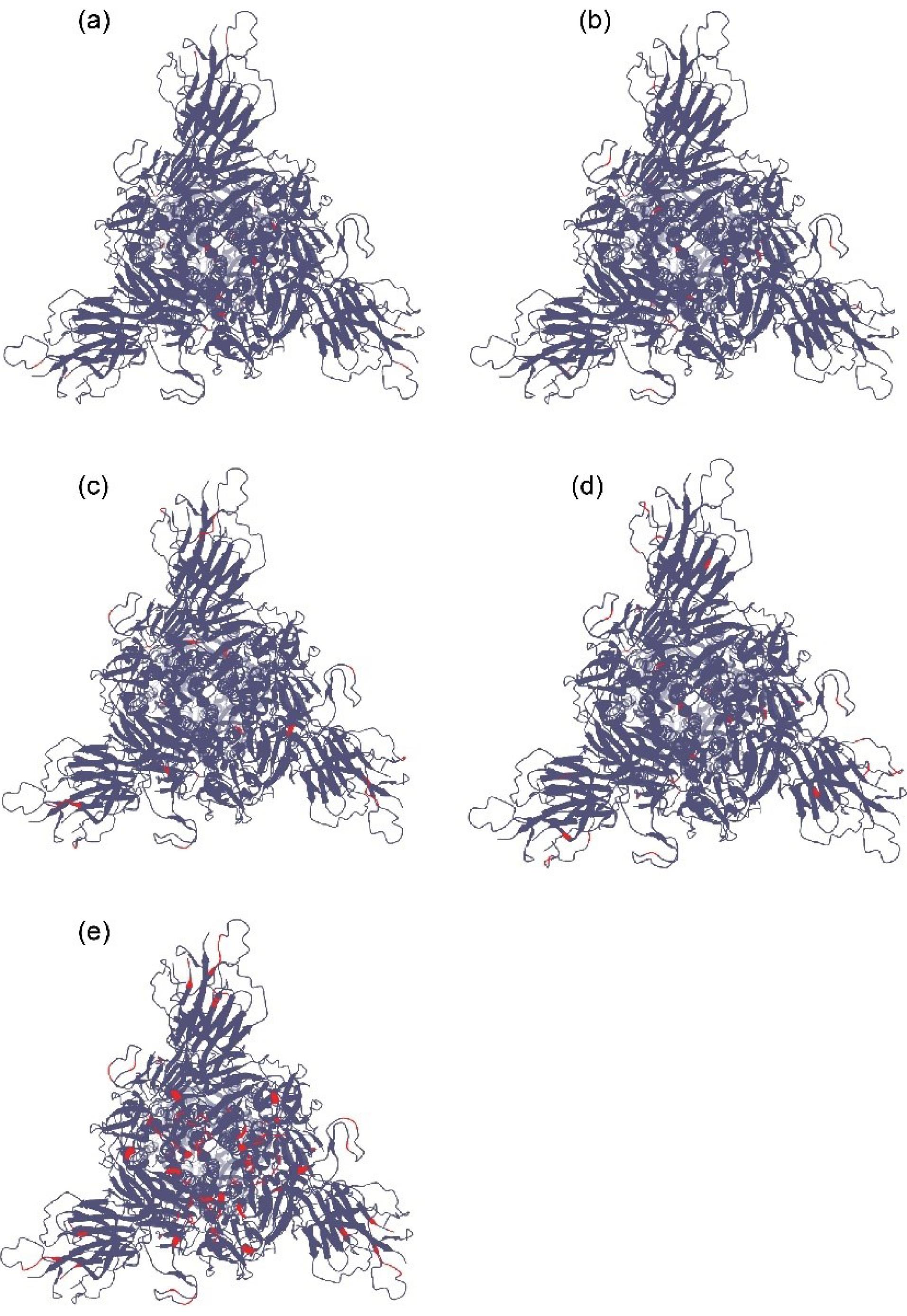
Figure 1.
(a) A 3-dimensional model of the Alpha variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (b) A 3-dimensional model of the Beta variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (c) A 3-dimensional model of the Delta variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (d) A 3-dimensional model of the Gamma variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (e) A 3-dimensional model of the Omicron variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1.20
.
(a) A 3-dimensional model of the Alpha variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (b) A 3-dimensional model of the Beta variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (c) A 3-dimensional model of the Delta variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (d) A 3-dimensional model of the Gamma variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1. (e) A 3-dimensional model of the Omicron variant of SARS-CoV-2 (S1 face). Red regions indicate mutated parts in comparison with Wuhan-Hu-1.20
In November 2021, a new variant was reported from South Africa, Omicron (B1.1.529) (Figure 1e), and was considered a new VOC. This new variant has rapidly spread worldwide and has become the dominant variant in many countries. Omicron has approximately 37 mutations in its spike protein in comparison with the initial Wuhan-Hu-1. It is thought that this variant was not derived from former strains and may have a different origin.18 The transmissibility rate of Omicron was much higher than that of other variants, and a higher peak was observed in the infection diagrams for each country.19
Types of mutations
In upper airway epithelial cells, the mutation N501Y enhanced the affinity of viral spike and replication. Based on the work of Liu et al, N501Y in the spike receptor-binding domain (RBD) is thought to lead to more contact with the human ACE2 receptor at K353, raising the receptor’s affinity. Using a bio-layer interferometry system, they conducted binding tests utilizing recombinant spike RBD and human ACE2 proteins to verify this. The binding was enhanced by the N501Y substitution. These findings suggest that N501Y increases viral fitness for replication in the upper airway, leading to enhanced transmission via enhanced spike receptor interactions. These results are in line with previous work showing that spike residue 501 is crucial for ACE2 receptor binding.21
The ability of serum-neutralizing antibodies to attach to the recombinant virus was affected by E484K. Sonia et al discovered that the E484K mutation reduced the neutralizing activity of human polyclonal sera produced in convalescent (infected with former strains) and vaccinated patients. These mutations are observed in SARS-CoV-2 varieties that belong to the B.1.351 and P.1 lineages, and they proposed that the single E484K mutation in the RBD influences the binding of serum polyclonal neutralizing antibodies from convalescent and vaccinated donors.22 Furthermore, Collier et al demonstrated that the recent emergence and dissemination of B.1.1.7 viruses carrying the spike E484K mutation causes a considerable loss of neutralization by BNT162b2 mRNA-elicited antibodies, convalescent sera, and mAbs.23
The combination of K417N and N501Y completely eliminated this antibody effect. The compensatory mechanism of Lys417Arg appeared to increase the S1 RBD-ACE2 free energy of binding. This may explain the greater spread of the virus in the UK and South Africa.24
New salt bridges and hydrogen bonds may occur as a result of mutations in BA.1.1 (K478), BA.2 (R400, R490, and R495), and BA.3 (R397 and H499). Mutations at Receptor-binding Motif (RBM) residues such as Q493R, N501Y, Q498, T478K, and Y505H in Omicron and sub-variants dramatically increased binding affinity with human ACE2. Relative to other mutations, such as K417N, interactions with Omicron variant mutations at residues 493, 496, 498, and 501 appear to restore ACE2 binding efficiency.25
Studies have shown that the D614G substitution may result in a higher spread rate in the sub-variant carrying it. Benton et al have shown that Asp614Gly contributes to promoting the opening conformation in the spike of the SARS-CoV-2 in contrast with wild-type, which facilitates the virus binding to ACE2.26
D614G did not affect the production, processing, or integration of S proteins into SARS-CoV-2 particles, as demonstrated by the study by Yurkovetskiy et al, although its affinity for ACE2 was lowered as a result of a higher dissociation rate. Cryo-electron microscopy analysis of the S protein trimer revealed that D614G breaks down interprotomer contact and shifts the structure toward an ACE2 binding-competent state, which is hypothesized to be a mechanism for virion membrane fusion with target cells. The neutralizing effectiveness of the antibodies directed against the S protein RBD was unaffected, which is compatible with this more open configuration.27
Liu et al reported that P681R mutation facilitates the virus entry to the cell through a cell-surface mediated entry pathway by enhancing cleavage of the whole spike to S1 and S2 parts. According to their findings, the Pro681Arg mutation is the primary mutation responsible for increasing S1/S2 cleavage and enhancing Delta variant replication.28
The P.1 lineage also includes a cluster of substitutions near the antigenic areas of the NTD, including L18F, which is known to decrease neutralization by certain antibodies, in addition to changes at positions 417, 484, and 501, which have already been reported.29 The NTD supersite also contains or is close to the substitutions T20N and P26S.30
Yamamoto et al highly recommended that the H655Y mutation of Omicron S is responsible for the altered usage of the entry pathways by assisting entry through the cathepsin B/L-dependent endosome pathway and impairing entry through metalloproteinase- and TMPRSS2-dependent cell surface pathways without changing the S1/S2 cleavage status.31
Each of these mutations can have specific effects, and their accumulation in a strain can directly affect the virulence of the virus. In Table 1, several important mutations can be seen.13
Table 1.
Mutation location and its effects on the virulence of SARS-CoV-2 VOC strains
16
|
Variant
|
Mutation
|
Location
|
Function
|
| B.1.1.7 (Alpha) |
N501Y |
RBD |
Increase the RBD affinity for binding to ACE2 receptor.
Increase the virus infectivity, transmissibility, and virulence. |
| P681H |
S1/S2 |
Facilitate the entry of the virus into the respiratory epithelial cells and increase its transmissibility |
| H69-V70del |
NTD |
Increase the virus infectivity |
| B.1.351 (Beta) |
N501Y |
RBD |
Increase the virus transmissibility |
| K417N, E484K |
RBD |
Increase the virus affinity for binding to receptors and escape from the immune system |
| L18F, D80A, D215G, R246I, LAL 242-244 del |
NTD |
Decrease the effect of neutralizing antibodies |
P.1 (B.1.1.28.1)
(Gamma) |
L18F, K417T, E484K, D614G |
S1 |
Escape from the immune system and reduce immunity due to antibodies.
Increase the transmissibility |
| B.1.617.2 (Delta) |
L452R |
RBD |
Increase the virus transmissibility and the affinity of binding to the ACE2 receptor.
Escape from the immune system and reduce immunity due to antibodies. |
| P681R |
Furin cleavage site |
| D614G |
S1 |
B.1.1.529
BA.1.1
BA.2
BA.3 (Omicron) |
N501Y |
RBD |
Increase the virus transmissibility
Increase the virus infectivity
Increase the virus pathogenicity |
| N679K, H655Y, P681H |
Furin cleavage site |
| R346K |
S1 |
| T19I, L24del, P25del, P26del, A27S, V213G, T376A, and R408S |
| R216del |
SARS-CoV-2 strain categorization based on effects of mutations
The major SARS-CoV-2 strains are divided into two categories by the WHO: strains that are variants of interest (VOI) and those that are variants of concern (VOC). VOC is a variant for which there is evidence of increased transmissibility, more severe illness (such as an increase in hospitalizations or deaths), a notable decrease in neutralization by antibodies produced during prior infection or vaccination, decreased efficacy of treatments or vaccines, or diagnostic detection failures. Variants of VOI strains have genetic markers linked to altered receptor binding, decreased neutralization by neutralizing antibodies, and a markedly elevated risk of transmission (Table 2).
Table 2.
Potential effects of mutations on the pathogenicity and transmissibility of SARS-CoV-2 variants.
4
|
|
Strains
|
Characteristics
|
| VOI |
B.1.525 (Eta)
B.1.526 (Lota)
B.1.617.1 (Kappa)
C.37 (Lambda)
B.1.621 (Mu) |
Changes in the receptor binding
Reduction of the neutralization of antibodies produced by previous covid-19 infection or vaccine injection
Reduction of the therapeutics
Potential diagnostic effect
The anticipable increase in the transmissibility and disease severity
Prevalence with a limited spread in the USA or other countries |
| VOC |
B.1.1.7 (Alpha)
B.1.351 (Beta)
P.1 (Gamma)
B.1.617.2 (Delta)
B.1.1.529 (Omicron) |
Increasing transmissibility
More severe disease
Significant reduction in neutralization by neutralizing antibodies due to vaccine injection or previous infection
Reduction in the effectiveness of therapeutics and vaccines
Error in diagnosis and vast cross-reaction in targets of diagnostic tests
Evidence for reducing vaccines effect in protection against severe disease |
Regarding the effects of vaccines against new SARS-CoV-2 variants, according to published data, the effectiveness of vaccines against mutant strains is thought to be reduced. Because most mutations occur in the S protein of the virus, and because most vaccines induce immune responses against the S protein, these responses have less effect on new mutations. The same is true for antibodies from previous infections.29,32,33
However, the extent of this effect varies depending on the vaccine used. Studies have shown that strains with the E484K mutation are more resistant to the effects of vaccine-induced neutralizing antibodies or in the serum of improved patients.22
Strain Alpha or B.1.1.7, was first documented in the United Kingdom by December 2020.12 According to studies, it is inferred that the Pfizer BioNTech vaccine has shown good performance against this virus34-36 Also based on the clinical results of the Oxford AstraZeneca vaccine, despite producing much fewer antibodies (nine folds less), the efficacy is 70.4%. This suggests that even with low levels of antibodies, it can provide good immunity against the virus.37
A beta variant, B.1.351, was first reported in South Africa in December 202013 and showed significant resistance to antibodies from Pfizer BioNTech and AstraZeneca vaccines. Mutations N501Y, E484K, and K417N/T significantly reduce the strength of antibodies against the virus, even in people who have been fully vaccinated.13 The neutralizing effect of antibodies produced by Pfizer BioNTech against this variant was reduced by up to 6.5-folds.38 The neutralizing effect of the antibodies against 1.351.B decreased by approximately 16-fold in comparison to B.1.1.7.16
In the Gamma or P.1 variant, which was reported from Brazil/Japan by Jan-2021 for the first time, due to more mutations in the spike region, it seems to be more resistant to existing vaccines than B.1.351. According to laboratory studies, the performance of these two strains against the Pfizer vaccine is almost the same.38,39 It can be understood that vaccines do not provide complete protection against these strains, but they are not entirely ineffective.
Regarding the Delta variant or B.1.617.2, which first emerged in India by Oct-2020, there was a noticeable decrease in the titer of neutralizing antibodies caused by Pfizer vaccine injection in the recipients in comparison with other variants 39. This reduction is approximately 2.5-fold in people receiving Pfizer and 4.3-fold in those receiving AstraZeneca.40 In addition, there was a three-fold reduction in the neutralizing antibody titers against B.1.617.2 relative to B.1.1.7.16
The situation is slightly different for the Omicron variant and B.1.1.529. It was first reported in South Africa and soon spread worldwide, causing the return of restrictions on COVID-19 in some countries. Omicron has more mutations than previous variants, leading WHO to place it in the VOC group.18 Based on clinical observations and its rapid dissemination, Omicron appeared to have a high rate of spread. However, it is not yet clear whether, like Delta, it will lead to an increase in ICU cases.
Sub-lineages of Omicron include BA.1, BA.2, BA.4, and BA.5; the last two, BA.4, and BA.5 become the dominant variants. These two strains have been reported to be more transmissible and resistant to immunity generated by prior variants. The new sub-lineages have more than 30 new mutations that alter the omicron’s pathogenicity, infectivity, and transmissibility due to alterations in the omicron spike structure in comparison with the primary strain of Omicron.25,41
Studies show that vaccine efficacy decreases 8-127 times due to these mutations as compared to the wild type of SARS-CoV-2, but the administration of booster shots improves this reduction to 12-35 folds. Studies have revealed that even after three doses of an mRNA vaccine, the efficiency of vaccine-induced immunity against Omicron is only approximately 66.3%, which is less than 85% against the Delta version. Additionally, the more recent subvariants BA.4 and BA.5, significantly evaded neutralizing antibodies produced by both infection and immunization.41
Reports from South Africa indicate that the rates of omicron hospitalization are lower than those of delta hospitalization. This risk of hospitalization was approximately 29% lower than that of the other variants. Because of mutations in the S1 region, it seems to be less sensitive to monoclonal neutralizing antibodies in the serum.42
An animal study on macaques has shown that injecting booster doses with a specific Omicron Moderna vaccine against the Omicron variant does not provide higher safety and protection than the Moderna vaccine against the primary covid-19 virus.43
As reported by the CDC, Omicron sublineages, including three variants BQ.1, BQ.1.1, and XBB.1.5, of which BQ.1 and BQ.1.1, have recently been dominant in the USA. CDC data showed that these variants spread relatively quickly. In addition, it has recently been reported that approximately 49 percent of circulating variants are XBB.1.5.44
Despite the variants containing N679K, H655Y, and P681H mutations in the furin cleavage region that appear more infectious,45 research has shown that these mutations weaken spike protein cleavage and reduce virus entry and proliferation in the alveolar cells of the lung that express TMPRSS2. This condition also causes weaker cell-cell connections in lung cells.46
The virus uses two pathways to enter the cells: the pathway that requires TMPRSS2 and ACE2, and the pathway that requires only ACE2. Therefore, Omicron species use the ACE2 pathway. TMPRSS2 is more abundant in the cells of the lower airways, but the Omicron spike is less able to bind to TMPRSS2 because of the mutations mentioned above, unlike the delta variant.46,47
In addition, the entry of Omicron into the cell was not inhibited by drugs targeting TMPRSS2. Studies on anti-Omicron antiviral therapies have shown that REGN2 cocktails have virtually no effect against Omicron in vitro, but other studies have shown that molnupiravir and remdesivir retain their effects.46
Some studies have reported the formation of new chemical bonds (salt bridges and hydrogen bonds) between RBD and ACE2 in the Omicron region, and these interactions compensate for the reduced binding affinity of RBD to ACE2 caused by mutations such as K417N. The omicron genome sequence analysis suggested that Omicron might not be derived from the currently circulating variants of SARS-CoV-2, and it might have a different origin.18,46
Studies on the protective efficacy of different vaccines showed that the Sinopharm vaccine demonstrated low efficacy against Omicron, even after the third shot. In a study, Yu et al reported that virus neutralization by the serum of Sinopharm vaccine recipients after the third dose was effective for the wild-type variant but decreased about 20 times against the Omicron variant.48
A large study from the United States found that during the predominance of the Omicron variant, the incidence of hospitalization for covid-19 in non-vaccinated individuals was 23 times higher than that in those fully vaccinated with booster doses. This amount was 5.3 times higher in comparison with vaccinated people without a booster dose. In addition, the incidence of COVID-19 in non-vaccinated individuals was 3.6 and 2 times higher than that in vaccinated individuals with and without the booster, respectively.49
Perez-Then E, reported that injection of one dose of Pfizer as a booster to individuals that had received two shots of inactivated vaccine CronaVac leads to increasing neutralizing antibody against Delta and ancestral variant which was similar to the response of individuals who received two shot mRNA vaccine.50
However, based on this article’s data, the previous booster (2x CronaVac vaccine) was unable to induce sufficient antibodies against Omicron; however, after taking one booster of Pfizer, it reached a detectable level in recipients.50
In addition, prior infection in individuals who received 2x CronaVac followed by 1x Pfizer as a booster had no effect on the amount of neutralizing antibodies against Omicron, whereas, in 2x mRNA recipients, prior infection elevated NA significantly.50
Ongoing research studies pertaining to COVID-19 and SARS-CoV-2 are currently underway, with the potential to modify our current understanding of the subject matter. As a result, it is important to acknowledge that the information presented in this article is subject to revision as new findings emerge.
Conclusion
Studies have shown that some mutations play an important role in escaping the virus from the immune system or neutralizing antibodies, more effectively bonding with host cells, or affecting the spreading rate of the virus. The consequences of these changes show themselves in reinfection shortly after individuals are cured, or also in a massive increase in patient numbers.
The results of these studies revealed that the effect of vaccines and neutralizing antibodies in people administered two doses of vaccine is significantly reduced against the Omicron variant, but vaccines are not completely ineffective. According to published reports, a third dose of the vaccine improves the titer of antibodies against this variant. Based on published reports, among the different vaccines evaluated, inactivated vaccines such as CoronaVac Sinopharm showed poorer performance than other vaccines even after a booster dose, whereas mRNA vaccines have shown higher efficiency in different trials. Also, as a study in China has stated that by prioritizing the administration of booster vaccines to high-risk individuals who have already been vaccinated, not only can their own protection be ensured, but also the potential transmission of the virus to other vulnerable groups can be effectively mitigated.51
In general, mutations alter virus behavior, and their accumulation in a variant may lead to different impacts on its transmissibility, virulence, immune escape, and resistance to vaccines, resulting in an increase in the number of patients and the severity of their illness; hence, permanent surveillance of virus genome mutations and new variants is very important in adopting important policies of governments and treatment systems to properly control the virus epidemic and to recognize and predict its possible behavior against vaccines and common treatment methods, as well as to improve these treatments and policies in the future.
Acknowledgments
The authors would like to thank the Biotechnology Research Center, Tabriz University of Medical Sciences, for supporting this work (Grant no. 68941).
Competing Interests
The authors declare that they have no conflicts of interest.
Consent for Publication
Not applicable.
Data Availability Statement
Not applicable.
Ethical approval
This study was approved by the Ethics Committee of Tabriz University of Medical Sciences (IR.TBZMED.REC.1401.008).
References
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020; 5(4):536-44. doi: 10.1038/s41564-020-0695-z [Crossref] [ Google Scholar]
- Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019; 17(3):181-92. doi: 10.1038/s41579-018-0118-9 [Crossref] [ Google Scholar]
- Zhang J, Xiao T, Cai Y, Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol 2021; 50:173-82. doi: 10.1016/j.coviro.2021.08.010 [Crossref] [ Google Scholar]
- Jahandar-Lashaki S, Farajnia S, Milani M, Hosseini MK, Yousefzadeh R. A novel multi-epitope vaccine against SARS-CoV-2 variants of concern strains applying immunoinformatics approaches. ImmunoAnalysis 2023; 3(1):7. doi: 10.34172/ia.2023.07 [Crossref] [ Google Scholar]
- Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res 2020; 178:104792. doi: 10.1016/j.antiviral.2020.104792 [Crossref] [ Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020; 11(1):1620. doi: 10.1038/s41467-020-15562-9 [Crossref] [ Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012; 4(6):1011-33. doi: 10.3390/v4061011 [Crossref] [ Google Scholar]
- Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 2017; 1(1):33-46. doi: 10.1002/gch2.1018 [Crossref] [ Google Scholar]
- O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 2021; 7(2):veab064. doi: 10.1093/ve/veab064 [Crossref] [ Google Scholar]
- O’Toole Á, Hill V, Pybus OG, Watts A, Bogoch II, Khan K. Tracking the international spread of SARS-CoV-2 lineages B117 and B1351/501Y-V2 with grinch. Wellcome Open Res 2021; 6:121. doi: 10.12688/wellcomeopenres.16661.2 [Crossref] [ Google Scholar]
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L. Assessing transmissibility of SARS-CoV-2 lineage B117 in England. Nature 2021; 593(7858):266-9. doi: 10.1038/s41586-021-03470-x [Crossref] [ Google Scholar]
- Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A. Emergence of SARS-CoV-2 B117 lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep 2021; 70(3):95-9. doi: 10.15585/mmwr.mm7003e2 [Crossref] [ Google Scholar]
- Ramesh S, Govindarajulu M, Parise RS, Neel L, Shankar T, Patel S. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines (Basel) 2021; 9(10):1195. doi: 10.3390/vaccines9101195 [Crossref] [ Google Scholar]
- Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY. Neutralization of variant under investigation B16171 with sera of BBV152 vaccinees. Clin Infect Dis 2022; 74(2):366-8. doi: 10.1093/cid/ciab411 [Crossref] [ Google Scholar]
- Hoffmann M, Hofmann-Winkler H, Krüger N, Kempf A, Nehlmeier I, Graichen L. SARS-CoV-2 variant B1617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep 2021; 36(3):109415. doi: 10.1016/j.celrep.2021.109415 [Crossref] [ Google Scholar]
- Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596(7871):276-80. doi: 10.1038/s41586-021-03777-9 [Crossref] [ Google Scholar]
- Fujino T, Nomoto H, Kutsuna S, Ujiie M, Suzuki T, Sato R. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg Infect Dis 2021; 27(4):1243-5. doi: 10.3201/eid2704.210138 [Crossref] [ Google Scholar]
- Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Tuttle KS. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022; 375(6582):760-4. doi: 10.1126/science.abn7760 [Crossref] [ Google Scholar]
- Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20(5):533-4. doi: 10.1016/s1473-3099(20)30120-1 [Crossref] [ Google Scholar]
- ViralZone. Sars-CoV-2 Circulating Variants. 2024. Available from: https://viralzone.expasy.org/9556.
- Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022; 602(7896):294-9. doi: 10.1038/s41586-021-04245-0 [Crossref] [ Google Scholar]
- Jangra S, Ye C, Rathnasinghe R, Stadlbauer D, Krammer F, Simon V. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2021; 2(7):e283-4. doi: 10.1016/s2666-5247(21)00068-9 [Crossref] [ Google Scholar]
- Collier DA, De Marco A, Ferreira IA, Meng B, Datir R, Walls AC, et al. SARS-CoV-2 B.1.1.7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies. medRxiv [Preprint]. February 15, 2021. Available from: https://pubmed.ncbi.nlm.nih.gov/33619509/.
- Fratev F. N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human-derived antibody: a free energy of perturbation retrospective study. J Chem Inf Model 2021; 61(12):6079-84. doi: 10.1021/acs.jcim.1c01242 [Crossref] [ Google Scholar]
- Kumar S, Karuppanan K, Subramaniam G. Omicron (BA1) and sub-variants (BA11, BA2, and BA3) of SARS-CoV-2 spike infectivity and pathogenicity: a comparative sequence and structural-based computational assessment. J Med Virol 2022; 94(10):4780-91. doi: 10.1002/jmv.27927 [Crossref] [ Google Scholar]
- Benton DJ, Wrobel AG, Roustan C, Borg A, Xu P, Martin SR. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc Natl Acad Sci U S A 2021; 118(9):e2022586118. doi: 10.1073/pnas.2022586118 [Crossref] [ Google Scholar]
- Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 2020;183(3):739-51.e8. 10.1016/j.cell.2020.09.032.
- Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep 2022; 39(7):110829. doi: 10.1016/j.celrep.2022.110829 [Crossref] [ Google Scholar]
- Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021; 592(7855):616-22. doi: 10.1038/s41586-021-03324-6 [Crossref] [ Google Scholar]
- McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021;184(9):2332-47.e16. 10.1016/j.cell.2021.03.028.
- Yamamoto M, Tomita K, Hirayama Y, Inoue JI, Kawaguchi Y, Gohda J. SARS-CoV-2 Omicron spike H655Y mutation is responsible for enhancement of the endosomal entry pathway and reduction of cell surface entry pathways. bioRxiv [Preprint]. March 21, 2022. Available from: https://www.biorxiv.org/content/10.1101/2022.03.21.485084v1.
- Greaney AJ, Loes AN, Crawford KH, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021;29(3):463-76.e6. 10.1016/j.chom.2021.02.003.
- Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B. SARS-CoV-2 501YV2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 2021; 27(4):622-5. doi: 10.1038/s41591-021-01285-x [Crossref] [ Google Scholar]
- Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y. Antibody resistance of SARS-CoV-2 variants B1351 and B117. Nature 2021; 593(7857):130-5. doi: 10.1038/s41586-021-03398-2 [Crossref] [ Google Scholar]
- Collier DA, De Marco A, Ferreira I, Meng B, Datir RP, Walls AC. Sensitivity of SARS-CoV-2 B117 to mRNA vaccine-elicited antibodies. Nature 2021; 593(7857):136-41. doi: 10.1038/s41586-021-03412-7 [Crossref] [ Google Scholar]
- Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv [Preprint]. January 25, 2021. Available from: https://pubmed.ncbi.nlm.nih.gov/33501442/.
- Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B117): an exploratory analysis of a randomised controlled trial. Lancet 2021; 397(10282):1351-62. doi: 10.1016/s0140-6736(21)00628-0 [Crossref] [ Google Scholar]
- Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 2021;29(5):747-51.e4. 10.1016/j.chom.2021.04.007.
- Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C. Neutralising antibody activity against SARS-CoV-2 VOCs B16172 and B1351 by BNT162b2 vaccination. Lancet 2021; 397(10292):2331-3. doi: 10.1016/s0140-6736(21)01290-3 [Crossref] [ Google Scholar]
- Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021;184(16):4220-36.e13. 10.1016/j.cell.2021.06.020.
- Shrestha LB, Foster C, Rawlinson W, Tedla N, Bull RA. Evolution of the SARS-CoV-2 Omicron variants BA1 to BA5: implications for immune escape and transmission. Rev Med Virol 2022; 32(5):e2381. doi: 10.1002/rmv.2381 [Crossref] [ Google Scholar]
- Ledford H. How severe are Omicron infections?. Nature 2021; 600(7890):577-8. doi: 10.1038/d41586-021-03794-8 [Crossref] [ Google Scholar]
- Gagne M, Moliva JI, Foulds KE, Andrew SF, Flynn BJ, Werner AP, et al. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits comparable B cell expansion, neutralizing antibodies and protection against Omicron. bioRxiv [Preprint]. February 4, 2022. Available from: https://www.biorxiv.org/content/10.1101/2022.02.03.479037v1.
- Centers for Disease Control and Prevention (CDC). COVID Data Tracker: Summary of Variant Surveillance. CDC; 2023.
- Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 2021; 6(7):899-909. doi: 10.1038/s41564-021-00908-w [Crossref] [ Google Scholar]
- Meng B, Ferreira IA, Abdullahi A, Saito A, Kimura I, Yamasoba D, et al. SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion. bioRxiv [Preprint]. December 22, 2021. Available from: https://www.biorxiv.org/content/10.1101/2021.12.17.473248v2.
- Madissoon E, Oliver AJ, Kleshchevnikov V, Wilbrey-Clark A, Polanski K, Orsi AR, et al. A spatial multi-omics atlas of the human lung reveals a novel immune cell survival niche. bioRxiv [Preprint]. November 27, 2021. Available from: https://www.biorxiv.org/content/10.1101/2021.11.26.470108v1.
- Yu X, Wei D, Xu W, Li Y, Li X, Zhang X, et al. Reduced sensitivity of SARS-CoV-2 Omicron variant to booster-enhanced neutralization. medRxiv [Preprint]. December 26, 2021. Available from: https://www.medrxiv.org/content/10.1101/2021.12.17.21267961v3.
- Danza P, Koo TH, Haddix M, Fisher R, Traub E, K OY. SARS-CoV-2 infection and hospitalization among adults aged ≥ 18 years, by vaccination status, before and during SARS-CoV-2 B11529 (Omicron) variant predominance - Los Angeles county, California, November 7, 2021-January 8, 2022. MMWR Morb Mortal Wkly Rep 2022; 71(5):177-81. doi: 10.15585/mmwr.mm7105e1 [Crossref] [ Google Scholar]
- Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, et al. Immunogenicity of heterologous BNT162b2 booster in fully vaccinated individuals with CoronaVac against SARS-CoV-2 variants Delta and Omicron: the Dominican Republic experience. medRxiv [Preprint]. December 29, 2021. Available from: https://www.medrxiv.org/content/10.1101/2021.12.27.21268459v1%20.
- Li K, Zhao Z, Wei H, Rui J, Huang J, Guo X. Feasibility of booster vaccination in high-risk populations for controlling coronavirus variants - China, 2021. China CDC Wkly 2021; 3(50):1071-4. doi: 10.46234/ccdcw2021.259 [Crossref] [ Google Scholar]