ImmunoAnalysis. 4:6.
doi: 10.34172/ia.4086
Original Article
c-Myc Inhibition Induces an Additive Effect with Cyclophosphamide in Acute Lymphoblastic Leukemia
Mohammad Sadeghi Conceptualization, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing, 1 
Maryam Panahi Data curation, Software, Validation, 2
Zahra Kheyrizadeh Data curation, Validation, 3
Leili Aghebati-Maleki Validation, Writing – review & editing, 2
Ali Akbar Movasaghpour Akbari Conceptualization, Supervision, 1
Abbas Ali Hosseinpour Feizi Supervision, 1
Sima Shahmohammadi Farid Supervision, 1
Sanam Dolati Formal analysis, Writing – review & editing, 1
Farhad Jadidi-Niaragh Funding acquisition, Supervision, Writing – original draft, Writing – review & editing, 2, 4, 5, * 
Author information:
1Hematology and Oncology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
2Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
3Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
4Department of Immunology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
5Research Center for Integrative Medicine in Aging, Aging Research Institute, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Background:
Cellular-myelocytomatosis (c-Myc), an oncoprotein and a transcription factor, is involved in several essential cellular processes. The c-Myc expression level is highly regulated in normal cells. It has been proved that c-Myc expression is deregulated in malignant cells due to rearrangements and mutations. The overexpression of this molecule is also reported to be present in acute lymphoblastic leukemia (ALL) as well, which is correlated with an unfavorable response to treatment, poor prognosis, and decreased overall survival. The upregulation of c-Myc results in increased proliferation, cell growth, and survival of ALL cells. Hence, making it an ideal target for leukemia treatment. This study evaluates the effect of c-Myc silencing combined with cyclophosphamide treatment, an FDA-approved chemotherapeutic.
Methods:
Peripheral blood and bone marrow samples (mononuclear cells) were derived from eleven ALL patients. To silence c-Myc, small interfering-RNA (siRNA)-lipofectamine was used. The efficacy of gene silencing was assessed by the qRT-PCR test. Next, the effect of c-Myc silencing combined with cyclophosphamide treatment in ALL primary cells was evaluated using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) test.
Results:
ALL cells were successfully transfected with c-Myc-siRNA. Also, treating cells with cyclophosphamide exerted a slight fall in the c-Myc mRNA level. The MTT test revealed that following the inhibition of c-Myc by siRNA, the viability of primary ALL cells decreased in response to cyclophosphamide treatment. Also, it was discovered that silencing c-Myc with siRNA combined with cyclophosphamide treatment significantly inhibits the growth of primary ALL cells compared to cyclophosphamide monotherapy.
Conclusion:
c-Myc possesses high potential in the treatment of several cancers. Our findings add ALL to this category as well. Silencing c-Myc sensitizes ALL cells to cyclophosphamide treatment and can help with the better treatment of the afflicted individuals.
Keywords: c-Myc, Cyclophosphamide, Acute lymphoblastic leukemia, siRNA
Copyright and License Information
© 2024 The Author(s).
This is an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Funding Statement
This study was funded by the Tabriz University of Medical Sciences (grant numbers: 66207 and 66392).
Introduction
Acute lymphoblastic leukemia (ALL), a very common childhood malignancy,1 results from the uncontrolled proliferation of lymphoid progenitors.2-4 Eighty-five percent of ALL cases account for B-cell lineage, while the rest affect T-cell progenitors.5-7 Several mutations are reported to be involved in the process of ALL development.8 ALL peaks at ages around 5 and 50.9 Pediatric therapeutic regimens include corticosteroids, asparaginase, alkaloids, and antimetabolites.10 Despite all the advances in chemotherapy strategies and recent improvements in response to treatments, only 30%-40% of adult ALL patients obtain long-term remission.11 Therefore, new therapeutic approaches are required for the successful and effective treatment of ALL.
Cyclophosphamide, an alkylating agent, widely used to treat different neoplasms,12 has been reported to possess immunosuppressive properties as well. Cyclophosphamide treatment combined with other chemotherapeutics to treat ALL is under study by several clinical studies. The agent is highly cytotoxic, therefore, immunotherapeutics-combined (which possess high specificity and less toxicity) cyclophosphamide treatment can reduce its required and effective dose 13. Nowadays, small interfering RNA (siRNA) is widely used in cancer research to evaluate the role of different genes in the process of cancer.14 SiRNAs induce the degradation of the target mRNA, therefore silencing the target gene. However, to safely deliver siRNAs into the cell, a vector/carrier is required.14,15
The MYC family genes comprise three distinct genes including MYC (c-MYC), MYCN, and MYCL. All three genes code for transcription factors with a helix-loop-helix leucine zipper structure.16 Cellular-myelocytomatosis (c-Myc), a crucial cell cycle regulator, is coded by the c- Myc gene, located on 8q24.17 c-Myc forms a heterodimer with Myc-Associated factor X (MAX) protein. The dimer is responsible for most of the c-Myc functions. C-Myc functions as a transcription factor downstream of several cellular signaling pathways, and therefore its expression level is highly regulated.18 C-Myc highly regulates cell cycle progression, cell death, nucleotide metabolism, and proliferation.19,20
The expression of c-Myc is reported to be dysregulated in various malignancies.20 Several hematological neoplasms have also been reported to show c-Myc aberrant expression, including Burkitt lymphoma (BL), diffuse large B cell lymphoma (DLBCL),21,22 and multiple myeloma (MM).23 Allen et al reported the c-Myc overexpression in B-ALL cells, which was correlated with poor prognosis, organomegaly, and elevated risk of long-term disease.19 Based on another study, B-ALL cells express c-Myc, which correlates with the expression of P53, mutations of TP53, and decreased overall survival.24 According to a case report, c-Myc rearrangement with Bcl-2 in a B-ALL patient severely weakened the patient’s prognosis. The patient presented with a significant increase in extra-nodal infiltration and disease progression, which ended up in no response to treatment and death.25
In the case of T-ALL patients, adults, in particular, the cure rate is only around 50% and only 7% of the afflicted people maintain overall survival after five years.26,27 Abnormal c-Myc expression keeps T-ALL leukemic stem cells active, and c-Myc silencing effectively stops leukemogenesis.28 C-Myc has been reported to be essential for T-ALL development as well.29,30 In addition, it has been found that through aberrant signaling of the NOTCH pathway, chromosomal translocations, and unknown mechanisms, c-Myc is upregulated and over-activated in T-ALL cells.31
According to a study, following the silencing of c-Myc, the viability of DLBCL cells in response to cyclophosphamide was decreased significantly, compared to cells only treated with cyclophosphamide 32. However, the results in the case of ALL are scarce. Therefore, our study is one of the leading and most novel studies in this field.
Considering the sufficient evidence on the role of c-Myc in ALL and its progression, as well as the effect of silencing it in other hematological malignancies, and due to the scarcity of data regarding ALL, this study aimed to investigate the effect of silencing c-Myc in combination with cyclophosphamide treatment. To our knowledge, this is one of the most pioneering studies evaluating the effect of this combination therapy on these cells.
Methods
Materials
Cyclophosphamide was supplied by Cayman Chemical Company. Human c-Myc gene targeting siRNA was bought from Santa Cruz Biotechnology, Inc. (catalog number: sc-29226). The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide MTT Assay Kit was supplied by the American Type Culture Collection (ATCC® 30-1010K).
Patient samples
Cellular samples were collected from eleven confirmed ALL patients at Shahid Ghazi Hospital, Tabriz, in accordance to the declaration of Helsinki. Patients’ demographic data are presented in Table 1.15 Using Ficoll PaqueTM Plus (GE Healthcare, Uppsala, Sweden) and a centrifugation system, mononuclear cells were separated from the whole blood according to the instructions provided by the manufacturer. Next, the separated cells were cultured in RPMI-1640 containing 20% FBS and 2% L-glutamine. Viable cells were counted prior to any downstream analysis.
Table 1.
Patients’ characteristics
|
Patients
|
Age (y)
|
Sex
|
WBC
(x103
/mL)
|
Platelet
(x103
/mL)
|
Hb
(g/dL)
|
LDH level
(Nl: Up to 480 U/L)
|
Hepatomegaly
|
Splenomegaly
|
Lymphadenopathy
|
C- ALL Ag
(CD10)
( % )
|
Subtype
|
| 1 |
3 |
F |
106.3 |
23 |
8.2 |
1801 |
Yes |
Yes |
Peripheral |
83 |
Pre B-cell |
| 2 |
11 |
M |
5.1 |
39 |
12.2 |
1372 |
Yes |
Yes |
No |
90 |
Pre B-cell |
| 3 |
4 |
M |
2.5 |
52 |
6.6 |
395 |
Yes |
Yes |
Peripheral |
23 |
Pre B-cell |
| 4 |
10 |
M |
8 |
141 |
10.3 |
360 |
Yes |
Yes |
Peripheral |
0 |
Pre B-cell |
| 5 |
8 |
M |
10.4 |
66 |
8.7 |
701 |
No |
Yes |
No |
92 |
Pre B-cell |
| 6 |
8 |
M |
4,02 |
74 |
9.8 |
1283 |
Yes |
Yes |
No |
0 |
Pre B-cell |
| 7 |
6 |
M |
9.9 |
8 |
2.4 |
375 |
NO |
Yes |
No |
79 |
Pre B-cell |
| 8 |
11 |
M |
6.5 |
54 |
9.8 |
2437 |
Yes |
No |
Peripheral |
0 |
Pre B-cell |
| 9 |
12 |
M |
74.2 |
21 |
9.9 |
1284 |
Yes |
Yes |
Mediastinal |
0 |
T-cell |
| 10 |
4 |
M |
41.3 |
53 |
11.5 |
1525 |
Yes |
Yes |
Mediastinal |
0 |
T-cell |
| 11 |
9 |
M |
56.4 |
57 |
10.1 |
1367 |
Yes |
Yes |
Mediastinal |
0 |
T-cell |
Abbreviation: WBC, white blood cells; LDH, lactate dehydrogenase; CALL Ag, Common ALL antigen; Hb, hemoglobin.
Cell transfection with siRNA
The cells were seeded at a 1 × 105 concentration per well in 96 well plates and were incubated for 24 hours at 37 °C. Next, lipofectamine 2000 (Invitrogen) was used to transfect cells with siRNA. C-Myc siRNA, control siRNA, and lipofectamine were diluted at recommended concentrations based on the instructions of the manufacturer with the opti-MEM medium. Subsequently, the compounds were mixed and incubated for 20 minutes at 24 °C. Finally, the cells were treated with the compounds containing 50 pM of siRNA for 24 or 48 hours.
Gene expression analysis
After the cell transfection with siRNA, a qRT-PCR test was run to ensure the efficient transfection of cells. First, using TRIzol Reagent (Invitrogen) the total RNA of ALL cells extracted. RNA was then transcribed into cDNA and stored at -20 ̊C. Next, SYBER Green PCR Master Mix (Thermo Fisher, US) was used to run a qRT-PCR with 1 μl of cDNA by a light-cycler 480 qRT-PCR system (Roche). The 2-ΔΔCT method was used to relatively measure the expression level of genes to β-actin mRNA level. The following primers were used: c-Myc Forward (F) primer: 5’-CCTGGTGCTCCATGAGGAGAC-3’, c-Myc Reverse (R) primer: 5’- CAGACTCTGACCTTTTGCCAGG -3’,33 β-actin F primer: 5’- CACCATTGGCAATGAGCGGTTC -3’, and β-actin R primer: 5’- AGGTCTTTGCGGATG TCCACGT -3’.34
Analysis of growth inhibition
To determine the Ic50 values for PBMCs and BMMCs, cells were cultured with increasing concentrations of the cyclophosphamide (5, 10, 15, 20, 25 μM). Subsequently, an MTT test was performed. The concentration that stopped cellular growth to 50% was calculated using GraphPad Prism V9.
To assess the effect of the combinational groups on the growth of cells, an MTT test was performed. Briefly, patient-derived cells were inoculated into a 96-well plate (1 × 104 cells/well) and incubated for 24 hours. Subsequently, cells were subjected to the following mixtures for 24 or 48 hours; untreated, lipofectamine, scramble siRNA, c-Myc siRNA, cyclophosphamide, lipofectamine-c-Myc siRNA, lipofectamine-c-Myc siRNA + cyclophosphamide, and DMSO (0.2%). 50 pM of siRNA and Ic50 of the drug were used in each group. Next, 10 μL of MTT reagent was added to each well and incubated for 4 hours. After incubating each well for 4 hours with 10 μL of MTT reagent, the medium was removed, and PBS was used to wash each well. A microplate reader (Thermo Fisher, Waltham, MA, USA) measured the absorbance of wells after 100 μL of DMSO was added to each well. Next, cell viability was calculated using this equation35:
Statistical analysis
The data were statistically analyzed by GraphPad Prism V9 and SPSS. Statistical significance was set at P < 0.05.
Results
Cells were efficiency transfected with siRNA using lipofectamine
To make sure that cells were efficiently transfected with siRNA, a qRT-PCR test was run.
We evaluated the effect of our treatments on c-Myc mRNA levels. Treating cells with lipofectamine-c-Myc siRNA significantly decreased the mRNA level of c-Myc, indicating the effective transfection of lipofectamine. The result was significant compared to the results of cells treated with lipofectamine alone, control siRNA, c-Myc siRNA without a vector, and untreated.
Also, we discovered that the c-Myc mRNA level was slightly decreased when cells were subjected to cyclophosphamide. The findings of the qRT-PCR test are presented in Figure 1.
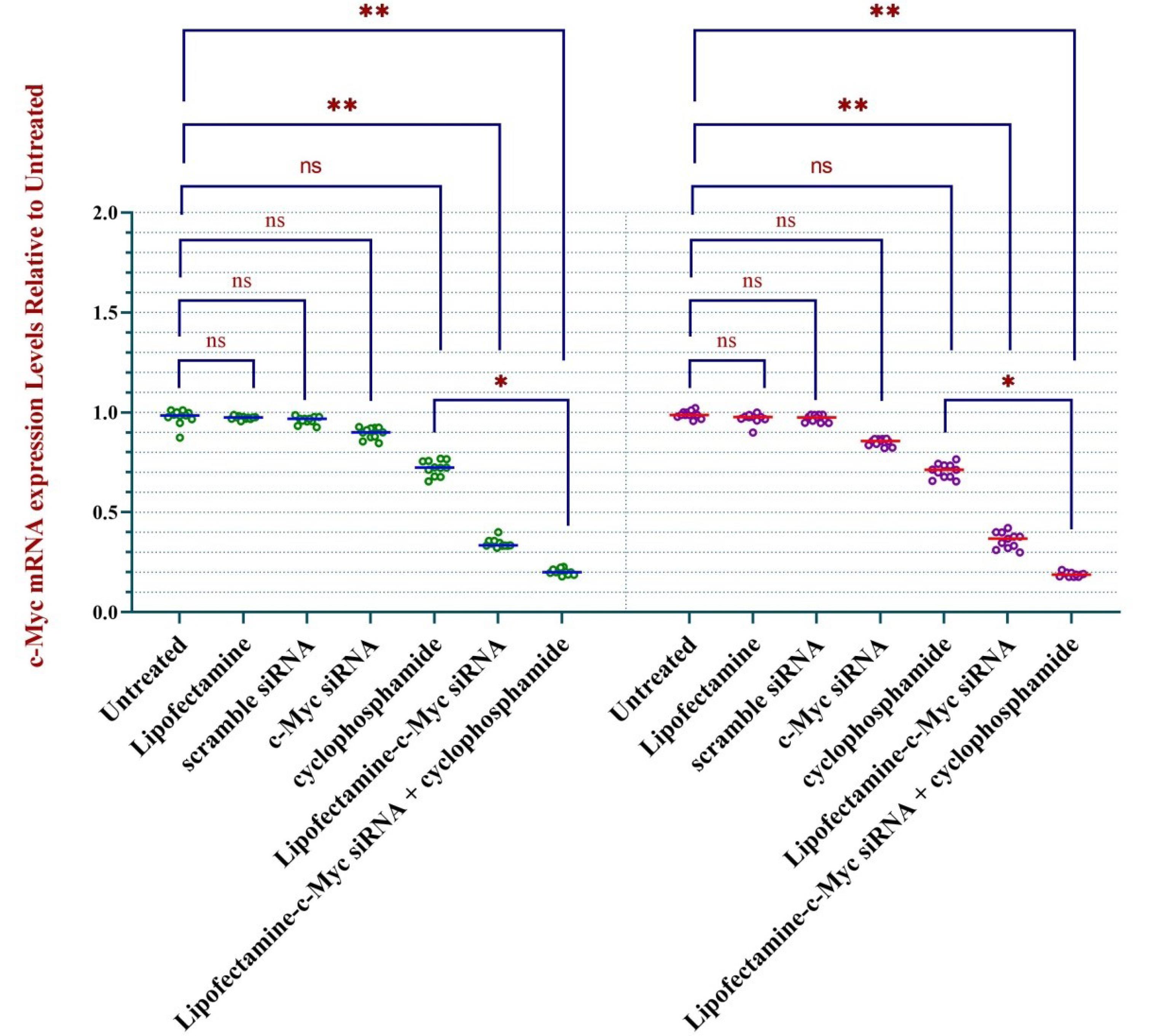
Figure 1.
Treating ALL cells with anti-c-Myc siRNA silences the c-Myc gene. Transfecting mononuclear cells separated from peripheral blood (PB) and bone marrow (BM) of ALL patients (n = 11) with siRNA using lipofectamine silenced c-Myc gene, as investigated using qRT-PCR. * represents p < 0.05 and ** indicates P < 0.01. Abbreviations: PBMC, peripheral marrow mononuclear cell; BMMC, bone marrow mononuclear cell; ns, non-significant
.
Treating ALL cells with anti-c-Myc siRNA silences the c-Myc gene. Transfecting mononuclear cells separated from peripheral blood (PB) and bone marrow (BM) of ALL patients (n = 11) with siRNA using lipofectamine silenced c-Myc gene, as investigated using qRT-PCR. * represents p < 0.05 and ** indicates P < 0.01. Abbreviations: PBMC, peripheral marrow mononuclear cell; BMMC, bone marrow mononuclear cell; ns, non-significant
The viability of cells in response to cyclophosphamide was decreased following the silencing of c- Myc
The IC50 = 22 μM for BMMCs and IC50 = 19 μM for PBMCs were measured by MTT tests following 24 hours incubation of cells with cyclophosphamide (Supplementary file 1, Figure S1).
Next, we aimed to evaluate the co-treatment effect of c-Myc silencing with cyclophosphamide on the growth of cells by running an MTT test after 24 or 48 hours incubation with the various treatments. The results are presented in Figures 2a and 2b.
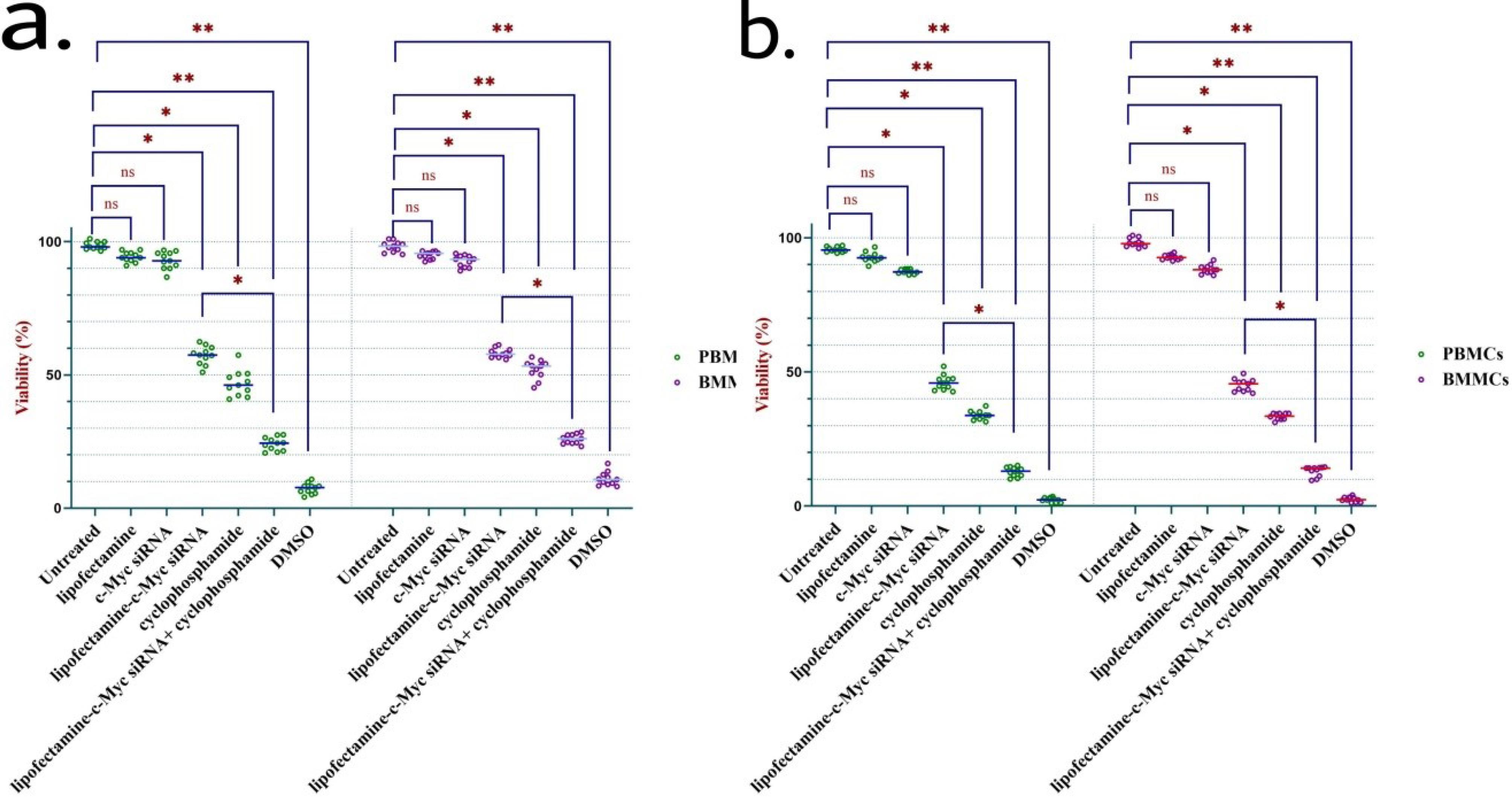
Figure 2.
c-Myc silencing decreased the viability of ALL cells to cyclophosphamide treatment. Silencing c- Myc in mononuclear cells separated from peripheral blood (PB) and bone marrow (BM) of ALL patients (n = 11) enhanced the cytotoxicity of cyclophosphamide in ALL cells following 24 hours (2a) and 48 hours (2b) incubation, determined by MTT test. * represents P < 0.05 and ** indicates P < 0.01. Abbreviations: PBMC, peripheral marrow mononuclear cell; BMMC, bone marrow mononuclear cell; ns, non-significant; DMSO, dimethyl sulfoxide
.
c-Myc silencing decreased the viability of ALL cells to cyclophosphamide treatment. Silencing c- Myc in mononuclear cells separated from peripheral blood (PB) and bone marrow (BM) of ALL patients (n = 11) enhanced the cytotoxicity of cyclophosphamide in ALL cells following 24 hours (2a) and 48 hours (2b) incubation, determined by MTT test. * represents P < 0.05 and ** indicates P < 0.01. Abbreviations: PBMC, peripheral marrow mononuclear cell; BMMC, bone marrow mononuclear cell; ns, non-significant; DMSO, dimethyl sulfoxide
We noticed that treating cells with lipofectamine-c-Myc siRNA, resulted in a considerable fall in viability, even though this effect was not as considerable as the effect of cyclophosphamide treatment. Next, the combined treatment effect was evaluated. The results indicated that cells with silenced c-Myc and treated with cyclophosphamide demonstrated the highest level of apoptosis compared to treatment with cyclophosphamide alone. The results were comparable to the controls, including untreated, treated cells with lipofectamine, and bare siRNA.
Therefore, inhibition of c-Myc sensitizes ALL primary cells to cyclophosphamide and helps with the better elimination of the neoplastic cells. Also, we found the treatment effect to be increased over the subsequent 24 hours of incubation.
Discussion
ALL, a neoplastic disease of the hematopoietic system, is characterized by the repletion of lymphoblasts in several organs.2,3 ALL in the elderly is associated with chemoresistance and therefore poor prognosis.11 Thus, new approaches to cancer therapy are required for the successful treatment of ALL. Interestingly, c-Myc is reported to be highly involved in cancer course.
C-Myc, an oncoprotein, and a transcriptional factor dimerizes with MAX to bind the DNA molecule and regulate gene expression.36 C-Myc is reported to induce cell growth, nucleotide and lipid synthesis, glucose metabolism, ribosome biogenesis, transcription, and translation of several genes.37 Since c-Myc is considered an oncogene, it is strictly regulated in healthy cells.38 Translocations, point mutations, and gene amplifications result in c-Myc deregulated expression which is reported in several types of neoplasms, hematopoietic malignancies in particular.18,39 In BL, t (8;14)(q24;q32) induces high levels of c-Myc which is involved in the cancer course.18,22 Many studies have suggested that c-Myc is upregulated in several cancers. T-ALL cells express high levels of c-Myc. It is proven that c-Myc expression is essential for the growth, and proliferation of T-ALL cells and the development of T-ALL.40 The overexpression of c-Myc has been proven in B-ALL cells as well. This upregulation is directly correlated with the mutations of the TP53 gene, unfavorable prognosis, and decreased overall survival of B-ALL patients.19,24,25 Therefore, we aimed to silence this gene with siRNA and study its impact on the viability of cells in response to an FDA-approved anticancer drug, cyclophosphamide.
To efficiently transfect cells with siRNA, lipofectamine was used. As seen in Figure 1, lipofectamine-c-Myc siRNA treatment significantly decreased the mRNA level of c-Myc. In addition, the c-Myc mRNA level was found to be slightly decreased in response to cyclophosphamide treatment. In the cell group receiving both siRNA and cyclophosphamide, the lowest level of c-Myc mRNA was noted. This was evident in both BMMCs and PBMCs. Next, the result of gene silencing on the growth of cells was evaluated by an MTT test. We found that silencing c-Myc sensitizes both T-ALL and B-ALL cells to cyclophosphamide treatment, as the rate of growth inhibition was considerably elevated in this group compared to cyclophosphamide monotherapy. Moreover, the effect of the treatments was further augmented over the next 24 hours incubation. Therefore, c-Myc inhibition possesses high potential in eliminating ALL cells.
Our findings are in support of several studies evaluating the role of c-Myc in cancer. Akyurek et al report the correlation between c-Myc rearrangements, decreased overall survival, and poor prognosis in DLBCL patients.41 According to Kendrick et al study, silencing c-Myc sensitizes DLBCL cells to cyclophosphamide treatment. Also, the combinational therapy induces a higher level of caspase-3 activity compared to cyclophosphamide monotherapy.32 Moreover, Skorski et al reported that following the inhibition of c- Myc, the proliferation of chronic myeloid leukemia (CML) cells was significantly decreased, which was comparable to the silencing of the BCR-ABL gene. The same research reports finding a synergism between BCR-ABL and c- Myc silencing. They added that the survival of CML mice xenograft models was improved following the silencing of the c- Myc gene.42,43 Therefore, not only c-Myc expression possess prognostic value in cancers but also synergizes with several anti-cancer drugs in eliminating neoplastic cells.
In this study, no significant difference was observed between patients’ samples in response to the treatments, which could be due to the scarcity of sample numbers. Therefore, future studies can examine the effect of this treatment in different subgroups of ALL as well as different samples of patients with larger statistical populations. In addition, the effect of this treatment can be studied with more advanced methods in leukemic stem cells to eliminate minimal residual disease, which was beyond the scope of this study. Also, studies can be conducted in vivo to measure survival following treatment.
Conclusion
To conclude, silencing c-Myc with siRNA decreases the viability of ALL cells. In addition, silencing c-Myc significantly sensitizes ALL cells to cyclophosphamide treatment. Future studies can confirm the existence of this additive effect between these two treatments with more samples and also investigate the effect of this combined treatment on different subtypes of ALL. Additionally, this data can help provide a new therapy approach for ALL patients with poor prognosis, due to its potential in improved elimination of ALL cells.
Acknowledgments
We appreciate the support of Tabriz University of Medical Sciences for this study (grant numbers: 66207 and 66392).
Competing Interests
The authors declare that they have no competing interests.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.
Ethical Approval
This study was approved by the ethics review board of Tabriz University of Medical Sciences (the ethical code: IR.TBZMED.REC.1399.256).
Supplementary File
Supplementary file contains Figure S1.
(pdf)
References
- Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin 2014; 64(2):83-103. doi: 10.3322/caac.21219 [Crossref] [ Google Scholar]
- Kato M, Manabe A. Treatment and biology of pediatric acute lymphoblastic leukemia. Pediatr Int 2018; 60(1):4-12. doi: 10.1111/ped.13457 [Crossref] [ Google Scholar]
- Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J 2017; 7(6):e577. doi: 10.1038/bcj.2017.53 [Crossref] [ Google Scholar]
- Nordlund J, Syvänen AC. Epigenetics in pediatric acute lymphoblastic leukemia. Semin Cancer Biol 2018; 51:129-38. doi: 10.1016/j.semcancer.2017.09.001 [Crossref] [ Google Scholar]
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009; 114(5):937-51. doi: 10.1182/blood-2009-03-209262 [Crossref] [ Google Scholar]
- Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J Clin Oncol 2012; 30(14):1663-9. doi: 10.1200/jco.2011.37.8018 [Crossref] [ Google Scholar]
- Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 2009; 360(26):2730-41. doi: 10.1056/NEJMoa0900386 [Crossref] [ Google Scholar]
- Bhojwani D, Yang JJ, Pui CH. Biology of childhood acute lymphoblastic leukemia. Pediatr Clin North Am 2015; 62(1):47-60. doi: 10.1016/j.pcl.2014.09.004 [Crossref] [ Google Scholar]
- Sant M, Allemani C, Tereanu C, De Angelis R, Capocaccia R, Visser O. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood 2010; 116(19):3724-34. doi: 10.1182/blood-2010-05-282632 [Crossref] [ Google Scholar]
- DeAngelo DJ, Jabbour E, Advani A. Recent advances in managing acute lymphoblastic leukemia. Am Soc Clin Oncol Educ Book 2020; 40:330-42. doi: 10.1200/edbk_280175 [Crossref] [ Google Scholar]
- Jabbour E, O’Brien S, Konopleva M, Kantarjian H. New insights into the pathophysiology and therapy of adult acute lymphoblastic leukemia. Cancer 2015; 121(15):2517-28. doi: 10.1002/cncr.29383 [Crossref] [ Google Scholar]
- Fraiser LH, Kanekal S, Kehrer JP. Cyclophosphamide toxicity Characterising and avoiding the problem. Drugs 1991; 42(5):781-95. doi: 10.2165/00003495-199142050-00005 [Crossref] [ Google Scholar]
- Hassan DS, Hasary HJ. Clinical implication of cyclophosphamide in oncology, hematology and bone marrow transplantation (BMT). Int J Innov Sci Res Technol 2022; 7(5):1073-9. [ Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL. Dynamic polyconjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci U S A 2007; 104(32):12982-7. doi: 10.1073/pnas.0703778104 [Crossref] [ Google Scholar]
- Hosseinpour Feizi AA, Vakili-Samiani S, Karpisheh V, Masjedi A, Izadi S, Adibfar S. Increased susceptibility to doxorubicin-induced cell death in acute lymphocytic leukemia cells by inhibiting serine/threonine WEE1 kinase expression using the chitosan-carboxymethyl dextran-polyethylene glycol-TAT nanoparticles. J Drug Deliv Sci Technol 2022; 77:103868. doi: 10.1016/j.jddst.2022.103868 [Crossref] [ Google Scholar]
- Adhikary S, Eilers M. Transcriptional regulation and transformation by MYC proteins. Nat Rev Mol Cell Biol 2005; 6(8):635-45. doi: 10.1038/nrm1703 [Crossref] [ Google Scholar]
- Shi W, Xu X, Huang R, Yu Q, Zhang P, Xie S. Plasma c-MYC level manifesting as an indicator in progression of breast cancer. Biomark Med 2019; 13(11):917-29. doi: 10.2217/bmm-2019-0073 [Crossref] [ Google Scholar]
- Dang CV. MYC on the path to cancer. Cell 2012; 149(1):22-35. doi: 10.1016/j.cell.2012.03.003 [Crossref] [ Google Scholar]
- Allen A, Gill K, Hoehn D, Sulis M, Bhagat G, Alobeid B. c-MYC protein expression in B-cell acute lymphoblastic leukemia, prognostic significance?. Leuk Res 2014; 38(9):1061-6. doi: 10.1016/j.leukres.2014.06.022 [Crossref] [ Google Scholar]
- Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N. Global regulation of nucleotide biosynthetic genes by c-MYC. PLoS One 2008; 3(7):e2722. doi: 10.1371/journal.pone.0002722 [Crossref] [ Google Scholar]
- Slack GW, Gascoyne RD. MYC and aggressive B-cell lymphomas. Adv Anat Pathol 2011; 18(3):219-28. doi: 10.1097/PAP.0b013e3182169948 [Crossref] [ Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-MYC onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A 1982; 79(24):7824-7. doi: 10.1073/pnas.79.24.7824 [Crossref] [ Google Scholar]
- Cobbold LC, Wilson LA, Sawicka K, King HA, Kondrashov AV, Spriggs KA. Upregulated c-MYC expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene 2010; 29(19):2884-91. doi: 10.1038/onc.2010.31 [Crossref] [ Google Scholar]
- Gao L, Harbaugh B, Parr K, Patel P, Golem S, Zhang D. MYC expression is associated with p53 expression and TP53 aberration and predicts poor overall survival in acute lymphoblastic leukemia/lymphoma. Am J Clin Pathol 2022; 157(1):119-29. doi: 10.1093/ajcp/aqab105 [Crossref] [ Google Scholar]
- Oberoi S, Dawson A, Marko D, Almiski M, Higgins R, Israels SJ. Precursor B-cell acute lymphoblastic leukemia with MYC and BCL2 rearrangements presenting as extensive extranodal disease in an adolescent. J Pediatr Hematol Oncol 2021; 43(4):e501-4. doi: 10.1097/mph.0000000000002051 [Crossref] [ Google Scholar]
- Fielding AK, Richards SM, Chopra R, Lazarus HM, Litzow MR, Buck G. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood 2007; 109(3):944-50. doi: 10.1182/blood-2006-05-018192 [Crossref] [ Google Scholar]
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet 2008; 371(9617):1030-43. doi: 10.1016/s0140-6736(08)60457-2 [Crossref] [ Google Scholar]
- King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 2013; 153(7):1552-66. doi: 10.1016/j.cell.2013.05.041 [Crossref] [ Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP. MYC-induced T cell leukemia in transgenic zebrafish. Science 2003; 299(5608):887-90. doi: 10.1126/science.1080280 [Crossref] [ Google Scholar]
- Roderick JE, Tesell J, Shultz LD, Brehm MA, Greiner DL, Harris MH. c-MYC inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood 2014; 123(7):1040-50. doi: 10.1182/blood-2013-08-522698 [Crossref] [ Google Scholar]
- Li Q, Pan S, Xie T, Liu H. MYC in T-cell acute lymphoblastic leukemia: functional implications and targeted strategies. Blood Sci 2021; 3(3):65-70. doi: 10.1097/bs9.0000000000000073 [Crossref] [ Google Scholar]
- Kendrick S, Muranyi A, Gokhale V, Hurley LH, Rimsza LM. Simultaneous drug targeting of the promoter MYC G-quadruplex and BCL2 i-motif in diffuse large B-cell lymphoma delays tumor growth. J Med Chem 2017; 60(15):6587-97. doi: 10.1021/acs.jmedchem.7b00298 [Crossref] [ Google Scholar]
- Marsoner F, Marcatili M, Karnavas T, Bottai D, D’Agostino A, Scarone S. Generation and characterization of an induced pluripotent stem cell (iPSC) line from a patient with clozapine-resistant Schizophrenia. Stem Cell Res 2016; 17(3):661-4. doi: 10.1016/j.scr.2016.11.005 [Crossref] [ Google Scholar]
- Kim TS, Shin YH, Lee HM, Kim JK, Choe JH, Jang JC. Ohmyungsamycins promote antimicrobial responses through autophagy activation via AMP-activated protein kinase pathway. Sci Rep 2017; 7(1):3431. doi: 10.1038/s41598-017-03477-3 [Crossref] [ Google Scholar]
- Joshi N, Hajizadeh F, Ansari Dezfouli E, Zekiy AO, Nabi Afjadi M, Mousavi SM. Silencing STAT3 enhances sensitivity of cancer cells to doxorubicin and inhibits tumor progression. Life Sci 2021; 275:119369. doi: 10.1016/j.lfs.2021.119369 [Crossref] [ Google Scholar]
- Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 2014; 4(1):a014357. doi: 10.1101/cshperspect.a014357 [Crossref] [ Google Scholar]
- Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov 2015; 5(10):1024-39. doi: 10.1158/2159-8290.cd-15-0507 [Crossref] [ Google Scholar]
- Levens D. You don’t muck with MYC. Genes Cancer 2010; 1(6):547-54. doi: 10.1177/1947601910377492 [Crossref] [ Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013; 45(10):1134-40. doi: 10.1038/ng.2760 [Crossref] [ Google Scholar]
- Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood 2017; 129(9):1124-33. doi: 10.1182/blood-2016-09-692582 [Crossref] [ Google Scholar]
- Akyurek N, Uner A, Benekli M, Barista I. Prognostic significance of MYC, BCL2, and BCL6 rearrangements in patients with diffuse large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone plus rituximab. Cancer 2012; 118(17):4173-83. doi: 10.1002/cncr.27396 [Crossref] [ Google Scholar]
- Skorski T, Nieborowska-Skorska M, Wlodarski P, Zon G, Iozzo RV, Calabretta B. Antisense oligodeoxynucleotide combination therapy of primary chronic myelogenous leukemia blast crisis in SCID mice. Blood 1996; 88(3):1005-12. [ Google Scholar]
- Skorski T, Nieborowska-Skorska M, Campbell K, Iozzo RV, Zon G, Darzynkiewicz Z. Leukemia treatment in severe combined immunodeficiency mice by antisense oligodeoxynucleotides targeting cooperating oncogenes. J Exp Med 1995; 182(6):1645-53. doi: 10.1084/jem.182.6.1645 [Crossref] [ Google Scholar]